Network clusters of viruses from this study (green) and the NCBI RefSeq database (light blue). Each node represents a viral genome/contig; the edge between nodes represents a significant relationship between the two connected viral genomes/contigs, with the shorter length representing a stronger connection strength. The details of the viral clusters and statistical results can be found in Supplementary Data 3. — Nature Geoscience
The Guliya Glacier, located over 6,000 meters high in the far northwest of the Tibetan Plateau, has for decades been one of the richest archives available to scientists to study large-scale paleoclimate changes. By analyzing ice core samples from the glacier, microbiologists have now reconstructed parts of the viral DNA remaining in it and identified nearly 1,700 virus species, about three-quarters of which are newly discovered.
Drilling into prehistoric ice has no health consequences for modern humans, as these long-dormant viruses likely infected other dominant microbes rather than animals or humans. However, researchers found that their adaptations significantly affected their hosts' ability to survive in extreme conditions during the fluctuations of Earth's climate cycles.
“Before this work, the connection between viruses and large-scale climate change on Earth was largely unexplored,” said ZhiPing Zhong, lead author of the study and a research associate at Ohio State University's Byrd Polar and Climate Research Center. “Glacial ice is so valuable and we often don't have the large amounts of material needed for virus and microbial research.”
As unprecedented warming continues to accelerate glacier melting, the race to collect these ice cores before they disappear forever has only increased their scientific value. For example, the ice layers studied in this work provided pristine snapshots of how viruses behaved during three cold-warm periods over the past 41,000 years.
The study was published today in Nature Geoscience.
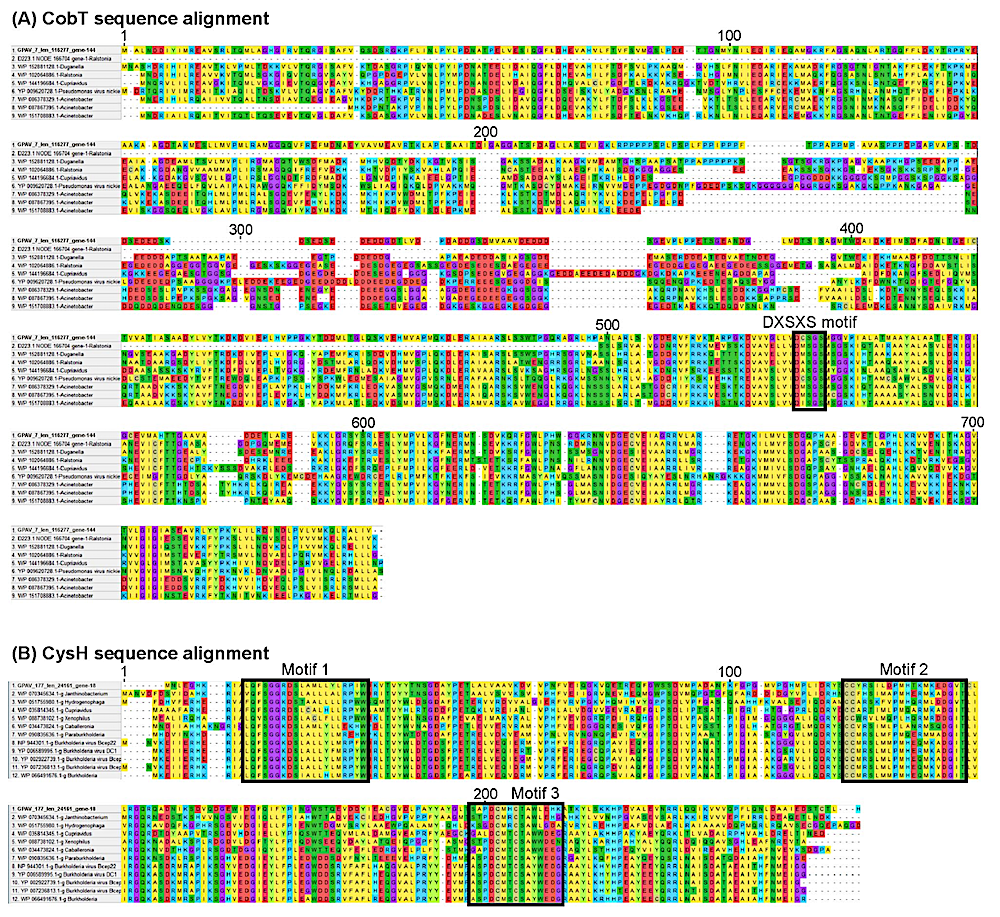
Multiple alignment of viral and microbial CobT (A) and CysH (B) protein sequences. The protein sequences of the AMGs and their most closely related bacterial-derived genes were included to illustrate the alignments and conserved motifs. The protein sequences were aligned using MAFFT (v.7.017) with the E-INS-I strategy for 1,000 iterations. The position numbers of the aligned sequences are indicated above. Conserved motifs were shown in black boxes. In the conserved motif “DXSXS” of the CobT sequences, “X” represents any amino acid. Possible conserved motifs 18 of the CysH sequences were identified using the alignment of all protein sequences in the phylogenetic analysis of Supplementary Figure S6B using the MEME26 tool (via default parameters). — Nature Geoscience
Of the various newly reported virus types, the most distinct viral community the team observed dates to about 11,500 years ago, when there was a major climate transition from the cold last ice age to the warm Holocene.
This suggests that microorganisms were responding to climate change as global temperatures changed from cool to warm, but it's too early to say for sure, Zhong said. “This at least suggests a possible link between viruses and climate change,” he said.
Using modern sequencing technologies, the team was able to examine the genetic signatures in more detail. The results also showed that while most of the viruses found in the glacier are unique to Guliya, about a quarter of the viruses overlap with known organisms from other parts of the world. “This means that some of them may have been introduced from areas such as the Middle East or even the Arctic,” Zhong said.
Taking the time to better understand how viruses evolved during periods of extreme climates can provide important insights into predicting how modern viruses are likely to respond to and interact with future ecosystem warming, researchers say. Additionally, since organisms found in ice cores expand the range of information researchers can gather about these time periods, finding and sequencing new stretches of ancient virus DNA could lead to an explosion of new puzzles and new insights.
“To me, this science is a new tool that can answer fundamental climate questions that we couldn't have answered otherwise,” said Lonnie Thompson, study co-author and professor of geosciences at Ohio State.
Refining these techniques on Earth will likely give scientists new tools to expand the search for life in space, aiding efforts to find microbes in the ice fields of Mars or beneath the icy shells of other celestial bodies, Thompson said.
Researchers seeking to make further links between viruses and climate here on Earth could also benefit from future technological advances as well as diverse scientific research approaches, the study says. But the clock is ticking, the authors say: These techniques must be implemented before warming threatens the glacial ice needed to preserve and continue to study Earth's rich history.
“I'm optimistic about what can be achieved here because if we work together, these techniques can help us address a wide range of scientific problems,” Thompson said.
Matthew Sullivan, co-author of the study and a professor of microbiology and civil, environmental and geodesy engineering at Ohio State University, said the study's success was due to how well the interdisciplinary approach of the Byrd Polar and Climate Research Center and the Ohio State University Center of Microbiome Science contributed to the development of new scientific knowledge.
“This type of opportunity brings together multiple disciplines, each with its own scientific language that hinders progress,” he said. “But the fact that we have been able to study ancient viruses and microbes in ice with this team is a testament to the support we have had in exploring new interfaces.”
The study was supported by the National Science Foundation, the Chinese Academy of Sciences, the Gordon and Betty Moore Foundation, the Heising-Simons Foundation, and the Joint Genome Institute of the U.S. Department of Energy.
Other co-authors include Ellen Mosley-Thompson, Olivier Zablocki, Yueh-Fen Li and Virginia Rich of Ohio State and James Van Etten of the University of Nebraska.
Glacier-preserved virus community on the Tibetan Plateau likely linked to warm-cold climate fluctuations, Nature Geoscience
Astrobiology,